oposSOM Browser of B-Cell Lymphoma
Help Index > oposSOM Browser of B-Cell Lymphoma
Live Demo
Use the search filed in the navigation bar and search for ‘SOM Analysis of 873 Malignant Lymphomas’, which leads to the general description of the data set. The link to the results browser can be found on the right part of the page.
1. Background
On molecular level cancers are complex and molecularly heterogeneous diseases. We present molecular landscapes of different cancer and disease
entities provided by our oposSOM analysis pipeline (Health Atlas entry). These landscapes enable discovery of
patterns of gene activation, their functional context, phenotype-associations and relations to alternative subtyping and corresponding prognosis.
2. Problem
As an example, we here present the expression and sample landscapes of B-cell lymphoma (850 cases collected in the German consortium
‘Molecular Mechanisms of Malignant B-cell Lymphomas; MMML). B-cell lymphoma is a very heterogeneous disease that decomposes into a multitude
of histo-pathological subtypes such as Burkitt’s lymphomas, diffuse large B-cell lymphomas, and follicular lymphomas. The expression and sample
landscapes provide an overview about clusters of concertedly regulated genes, which we, in turn, used to refine the subtype classification scheme.
Understanding of cancer biology and treatment represents a complex problem relating to gene regulation and dysfunction of cellular processes, and it
requires comprehensive tools that allow navigation in the data, their visualization in an intuitive interactive way and, linking to supporting information.
3. Solution
The oposSOM Browser provides this interactive functionality based on a series of bioinformatics tools implemented in the oposSOM-pipeline,
which is available as R-package in the Bioconductor repository and on the Health Atlas. The beta-version of the Browser demonstrates basic functionalities
that will be extended by a series of novel functionalities and other disease entities.
4. Guided Tour
I) Data portraying (Demo-Link)
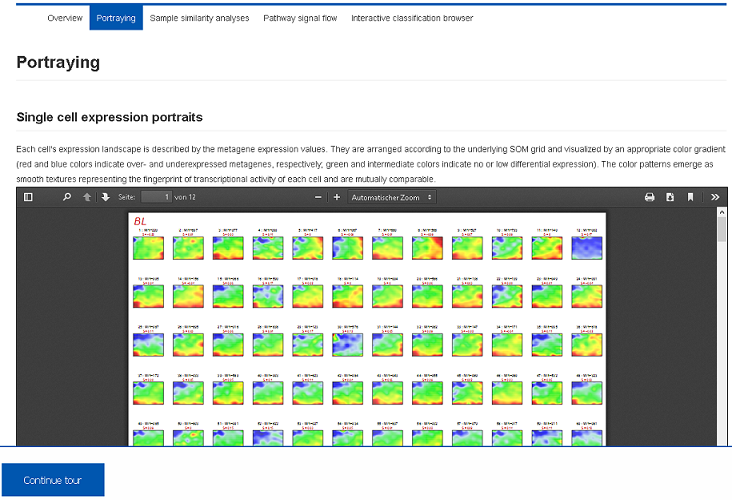
For each patient the SOM-method provides a ‘personalized’ portrait of the expression landscape serving as fingerprints of transcriptional activity of the samples. From these fingerprints, co-regulated gene modules are extracted and characterized. Report PDF files are stored in the Health Atlas and can be downloaded or previewed using the browser.II) Sample diversity analysis (Demo-Link)
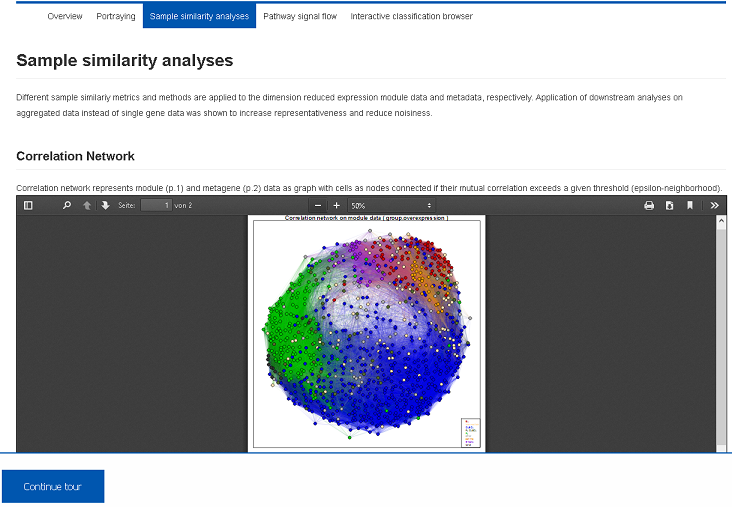
Diversity analysis of the samples is performed using multiple complementary approaches such as correlation networks, hierarchical clustering heatmaps and component analyses. They are applied to detect molecularly distinct subgroups of the samples, to detect outlier samples and to visualize transcriptional diversity of the data set. Report PDF files are stored in the Health Atlas and can be downloaded or previewed using the browser.III) Functional mining (Demo-Link)
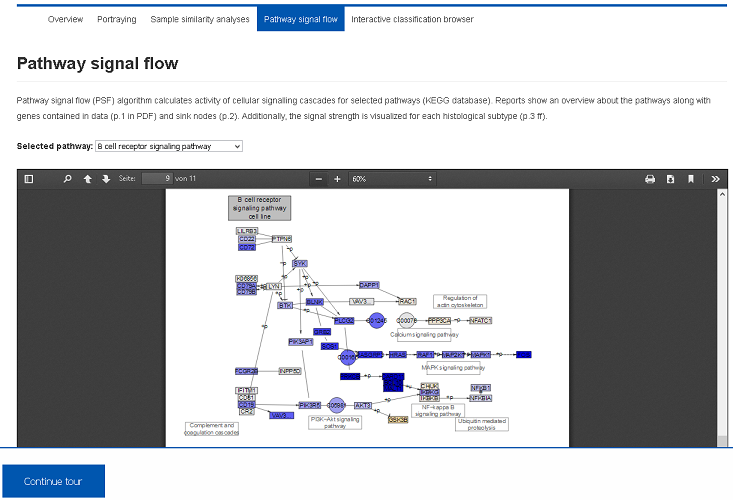
We applied Pathway Signal Flow (PSF) algorithm to calculate activity of cellular signalling. Importantly, this approach explicitly involves pathway topologies and provides proper visualization of the signal strengths. Several pathways can be inspected using the select box.IV) Alternative classifications (Demo-Link)
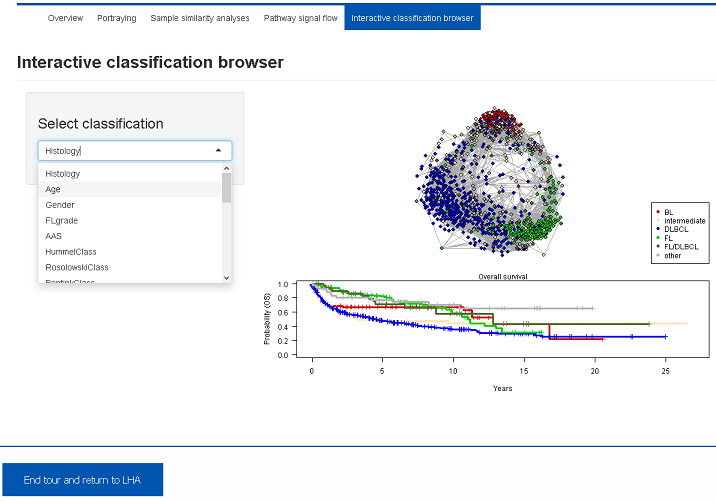
Several molecular, clinical and phenotypical classifications were applied to this data set. This Shiny app visualizes distribution of the subgroups in the sample correlation network and provides the corresponding survival curves. Several alternative classifications can be inspected using the select box.5. Results and Outlook
The oposSOM browser provides an intuitive and easy-to-use tool for discovering molecular data of selected diseases. The beta-version will be
extended into a comprising ‘cancer query tool’ which allows for direct comparison of different disease entities in terms of common molecular and
functional patterns.